Add a 2D peak as a 2D compound to the processing method
The expected compounds are stored in a compound table as part of the processing method. The following procedure describes how you add peaks as new 2D compounds to the compound table.
Preparations
|
From the Contour Plot 2D
-
Load a single sample or result set with 2D data that has a 2D-LC method linked to it.
In the Contour Plot 2D window, click
 Show 2D peak regions.
Show 2D peak regions.The regions that the software recognizes as one 2D peak are surrounded by a border.
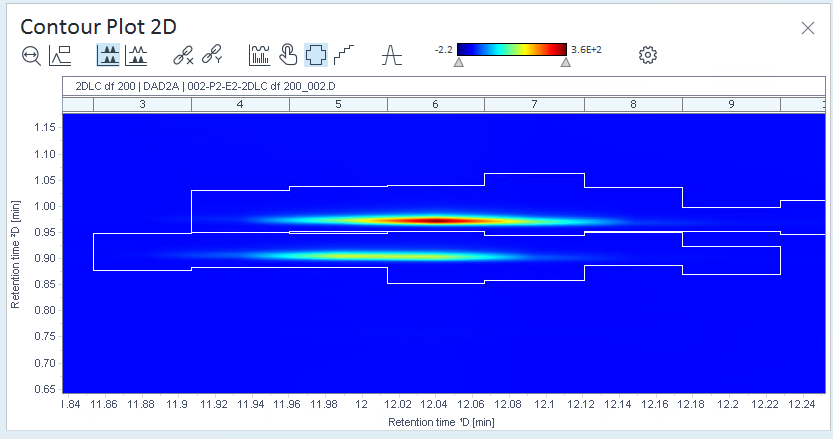
If the peak regions do not correspond to your compounds,optimize the integration parameters, optimize the method, or integrate manually.
-
In the contour plot, select the 2D peak region you want to add.
To select multiple 2D peak regions, hold the Ctrl key while clicking. To select a range of 2D peak regions, hold the Shift key while clicking.
-
Right-click the selected 2D peak region, and select Add 2D peak as 2D compound to method (or Add multiple 2D peaks as 2D compound to method respectively, if you selected multiple 2D peak regions).
Alternatively, add the 2D compound for all ²D signals of the linked data:
NOTE: Ensure that all signals are aligned (see Align signals over two dimensions). This is required because the same expected retention time will be set for all signals.
Right-click the selected peak, and choose Add 2D peak as 2D compound to method for all signals (or Add multiple 2D peaks as 2D compound to method for all signals respectively, if you selected multiple 2D peak regions).
Adjust the compound parameters as required. See Compound identification parameters.
From the Chromatograms ²D
-
Load a single sample or result set with 2D data that has a 2D-LC method linked to it.
-
In the Chromatograms ²D window, select the peak that you want to add.
-
Right-click the selected peak, and choose Add 2D peak as 2D compound to method from the context menu.
Alternatively, choose Add 2D peak as 2D compound to method for all signals to create a 2D compound for each ²D signal.
A 2D compound is added to the compound table of the selected method. If you created the 2D compound for all signals, a copy of the selected 2D compound is created for each ²D signal present in the data that is linked to the method.
By default, the compound parameters are set as follows:
Name: peak@RT1D,RT2D, where RT1D is the retention time of the first dimension detector and RT2D is the retention time of the second dimension detector
Signal: the ²D signal from which you selected the peak
Exp. ¹D RT: the expected retention time of the selected peak in the first dimension
Absolute RT Window ¹D (min): the window in minutes around the expected retention time of the peak in the first dimension (set to 0)
Relative RT Window ¹D (%): the window in percent (%) around the expected retention time of the peak in the first dimension (set to 1)
Exp. ²D RT: the expected retention time of the selected peak in the second dimension
Absolute RT Window ²D (min): the window in minutes around the expected retention time of the peak in the second dimension (set to 0)
Relative RT Window ²D (%): the window in percent (%) around the expected retention time of the peak in the second dimension (set to 1)
Adjust the compound parameters as required. See Compound identification parameters.
See Also
base-id: 10237449739
id: 10237449739